3D-Nanoprinted Microchannel Plate (3DN-MCP) Detector Functionalized by Atomic Layer Deposition (ALD) for Mass Spectrometry of High-Mass Proteins
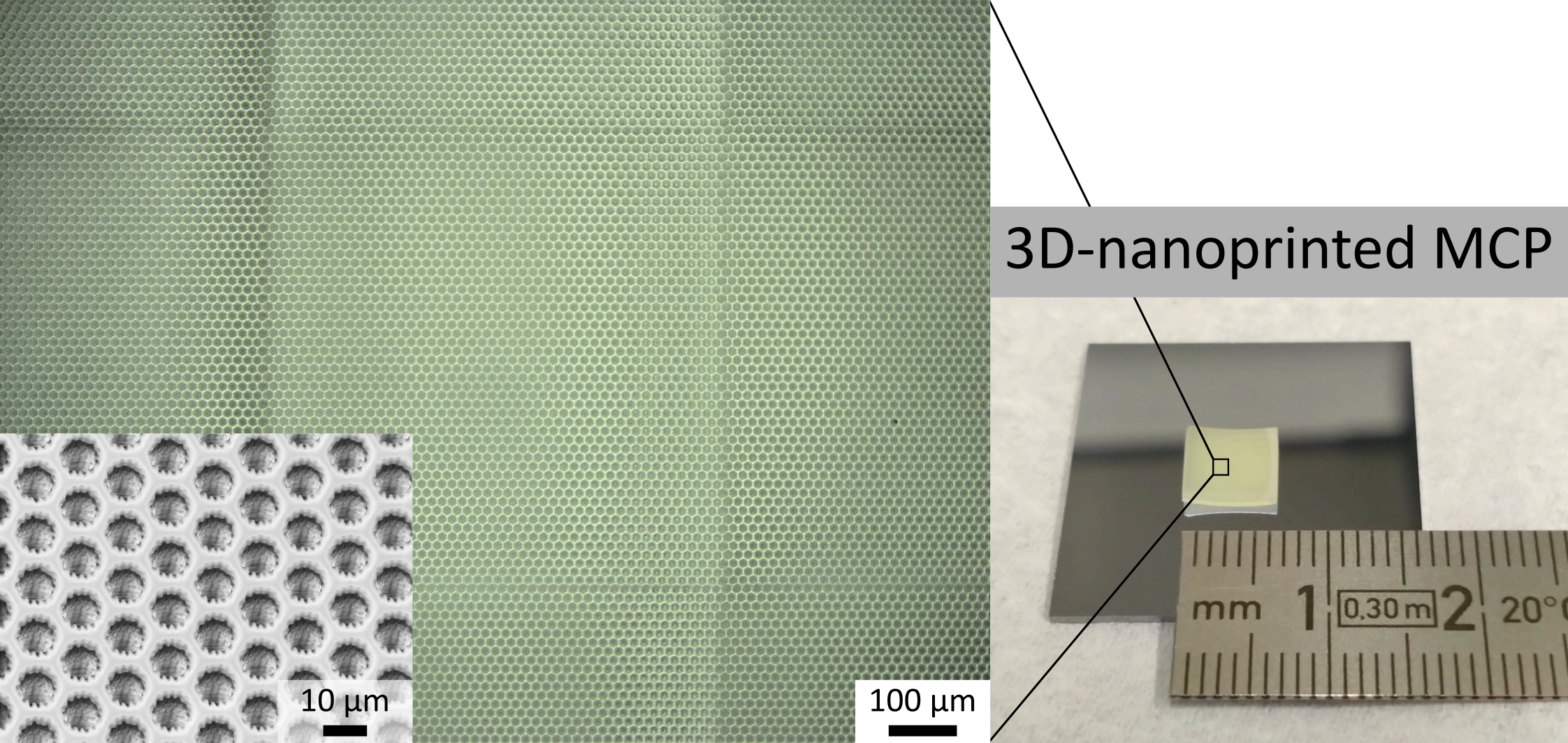
The investigation of intact proteins is a key aspect of top-down proteomics that reduces sample complexity and preserves information on modifications at the protein level, which will ultimately help to advance the understanding of complex disease mechanisms. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry allows for generating such low-charge-state ions from intact proteins. Typically, microchannel plates (MCPs) are used as detectors for MALDI-TOF mass spectrometry—whose detection mechanism relies on the release of secondary electrons upon ion impact—as they offer fast response times and high spatial resolution. However, the detection efficiency of conventional MCPs decreases considerably with increasing ion mass, preventing the sensing of intact high-mass proteins. The goal of this project is to explore the design freedom offered by 3D-nanoprinting (3DN) using two-photon polymerization for the generation of MCP structures beyond the classical straight-channel shape, which is dictated by the conventional fabrication process of glass MCPs. Subsequently, optimization of the 3DN-MCP’s gain will be carried out by precisely tuning the electrical properties of the emissive and the resistive layer deposited by atomic layer deposition (ALD), to achieve a high secondary electron yield and allow for the fast replenishment of electrons, respectively. Furthermore, a concept for integrating a picosecond infrared laser (PIRL) type system will be prepared to allow for the soft ablation of intact biomolecules replacing the conventional MALDI ion source. The combination of a tailor-made, ALD-functionalized 3DN-MCP detector with the future modification of the ion source may pave the way for the high-throughput investigation of intact high-mass proteins by mass spectrometry.




